|
Size: 17565
Comment:
|
← Revision 133 as of 2013-12-03 20:48:34 ⇥
Size: 17601
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 5: | Line 5: |
| = The Definition of MicroRNA = | = The Definition of MicroRNA and the Principles of its Operation = |
Itt írjon a(z) MicroRNA-ról/ről
The Definition of MicroRNA and the Principles of its Operation
Definition
MicroRNAs are a recently identified class of cellular RNAs that function as post-transcriptional regulators of protein expression in both plants and animals, although with some differences in the mechanism of function. The active, mature miRNAs are 17–24 base, single-stranded RNA molecules expressed in eukaryotic cells and are known to affect the translation or stability of target messenger RNAs. Each miRNA is believed to regulate multiple genes, with predictions that greater than one third of all human genes may be regulated by miRNA molecules. The explanation of this discovery is the fact that a normal miRNA exhibits 100 target sites and therefore regulates a major fraction of the protein-coding genes. These target sites can be grouped into two broad categories which are; the 59 dominant sites having complementarity to the miRNA 59 end and functioning with little or no support from pairing to the miRNA 39 end. In contrast, 39 compensatory sites have insufficient 59 pairing and require strong 39 pairing for function (Lewis et al, 2005).
(J.Brennecke et al, 2005 )
Figure 1: Three classes of miRNA target sites
History
In the early 1990’s, the genetic material of a particular species, either in the Plant or Animal Kingdom, was considered to be the regulator of cellular processes by following a basic formula: DNA synthesizes RNA, RNA itself makes proteins, and proteins are the major factors carrying out all crucial processes in the cells. Recent research, however, revealed the existence of a new world of “small RNAs” that do not synthesize proteins but rather control the protein coding RNAs (Steven Buckingham 2003). In 1993 Victor Ambros, Rosalinda Lee and Rhonda Feinbaum discovered miRNAs during a study of the gene lin-14 in Caenorhabditis elegans nematode development. The very first miRNA to be discovered was the lin-4 encoded by the lin-4 gene. This first miRNA was responsible for the regulation of the lin-4 protein abundance in the C. elegans nematodes. Particularly, 61- nucleotide precursor of the lin-4 gene matured into a 22-nucleotide RNA that showed a partial complementarity in certain sequences with multiple sequences in the three prime untranslated region of the mRNA (3’UTR). This complementarity was sufficient to inhibit the translation of the lin-14 messengerRNA into the lin-14 protein. The second miRNA to be identified in 2000 was the let-7 which has been verified in many species (Lewis et al, 2005).
(Marek Mraz, 2004)
Figure 2: Traditional mRNA
miRNA plays a significant role in many cellular processes including;
- Post-transcriptional regulation of gene expression
- Regulation of cell proliferation and apoptosis processes which are important in cancer formation.
- Importance in different kinds of development e.g. heart’s development.
- Differentiation of hematopoietic stem cells
- Embryogenesis and differentiation
- Metabolic regulation
- Virus Resistance
Plants Vs Animals
Research clearly revealed that miRNA plays a crucial role in all types of eukaryotic organisms in the Plant and Animal Kingdom. However, miRNA has evolved differently in these two kingdoms with diverse types of functions. In the Animal Kingdom, miRNAs predominantly regulate gene expression whereas in the Plant Kingdom they act to inhibit target gene elimination. A significance difference between plants and animals’ miRNAS is in target recognition. In plants, miRNA base sequence has near perfect complementarity to it's target site, thus resulting in a direct cleavage of the miRNA’s target site. In addition, binding of miRNAs’ target site occurs in both coding and untranslated regions. In contrast, animals have a completely different mechanism where the protein repression occurs with translation, inhibition as well as miRNA degradation. Moreover, in animals a partial complementarity exists regarding their target sites while the miRNA target recognition is driven through the canonical 6-8 nucleotide “seed sequence” found at the 5’ end of an animal’s mRNA (Marek Mraz, 2004).
Biogenesis
microRNA Transcription
Most miRNA genes are found in intergenic regions or in oriented antisense to neighbouring genes and therefore contain their own miRNA gene promoter and regulatory units. As much as 40 percent are said to lie in the introns of protein and non-protein coding genes and rarely in exons. In the nucleus, Polymerase II (POL II) is usually used to transcribe miRNA by encoding parts of the genome, often through binding to a promoter found near the sequence destined to be the hairpin loop of the pre-miRNA. This produces a transcript that is capped at the 5’ end, polyadenylated with multiple adenosines to give a (poly)A tail and spliced to form pri-miRNA. Pol II produces the mRNAs and some noncoding RNAs, including the small nucleolar RNAs (snoRNAs) and four of the small nuclear RNAs (snRNAs) of the spliceosome.The general transcription factors interact with promoter sequences at the 5’ end of genes and assist Pol II in initiating transcription. Once initiated, Pol II continues to transcribe by elongating the nascent transcript one nucleotide at a time in the 5’ to 3’ direction. At the 3’ end of the gene Pol II terminates transcription and releases the completed transcript (David P. Bartel, 2004).
Nuclear Processing
The miRNAs may be produced from their own genes or from introns. All miRNAs mature through a series of processing steps. The miRNAs are synthesized in cells as long primary transcripts (pri-miRNAs) that often contain thousands of nucleotides. The first step is the nuclear cleavage of the pri-miRNA, which liberates a ∼60–70 nt stem loop intermediate, known as the miRNA precursor, or the pre-miRNA. This processing is performed by the Drosha Ribonuclease endonuclease(RNase III) along with an RNA-binding protein DGCR8 or Pascha (partner of Drosha) which cleaves both strands of the stem at sites near the base of the primary stem loop producing pre-miRNA. Pre-miRNAs are typically 60–100 nt in length and serve as the substrate for nuclear export. Jointly, Drosha and DGCR8 form the microprocessor complex. Drosha cleaves the RNA duplex with a staggered cut typical of RNase III endonucleases, and thus the base of the pre-miRNA stem loop has a 5’ phosphate and ∼2 nt 3’ overhang. In general, the majority of the the microRNAs that are located within introns are cleaved by Drosha. However, via deep sequencing of D. melanogaster and C. elegans RNAs allowed the identification of a novel class of intronic miRNAs that do not contain the 10-bp helix at the base of the miRNA hairpin normally required for Drosha cleavage. Mirtrons are pre-miRNAs that can be produced without having to undergo the microprocessor machinery if they are directly spliced from the introns in which they reside. These mirtrons have traditionally been thought to only exist in drosphila. The mirtrons turned out to be processed directly by a spliceosome, instead of Drosha. Both Drosha-processed pre-miRNAs and mirtrons are processed by Dicer. In comparision to other introns, mirtrons are flanked by 5′ and 3′ splicing sites, and contain branch point sequences. Unlike the pre-miRNAs generated by Drosha, the mirtrons generated by the spliceosome need to be linearized by an enzyme and folded into hairpins, prior to exportation to the cytoplasm by Exportin-5. This hairpin structure is exported from the nucleus once the nuclear membrane protein Exportin-5 recognises the 2 nucleotide overhang on the 3 end of the pre-miRNA. Exportin-5 transports it into the cytoplasm using ran-guanine triphosphatase (Ran-GTP) (Michael Faller et al, 2008).
Nuclear Transport
The nuclear transport of miRNA occurs through nuclear pores which are embedded on the nuclear envelope. Soluble compounds are needed to transport the molecules across the nuclear membrane. The receptor of the soluble transport compounds binds to specific sequences of the substrate and escorts them through the nuclear pores by interacting with proteins termed nucleoporins. The receptors which are involved in transport belong to nuclear transport receptor family in which importin β and transportin are the key representatives. However, it has been recently shown that exportin 5 may be the only receptor responsible for the transport of pre-miRNA. As pre-miRNA reaches the cytoplasm, it will be further cleaved to mature miRNAs (Kim V. Narry, 2004).
Cytoplasmic Processing
In the cytoplasm an RNase III enzyme named Dicer is present. Dicer cleaves the pre-miRNA into shorter segments of 20-25 base pairs. This enzyme acts by unwinding the duplex to accomodate it's attachment to one of the strands. The strand selected is more thermodynamically unstable and has a weaker base pairing. The second strand called passenger strand becomes degraded. The resulting product of this process is the RNA induced silencing complex which is capable of degrading messenger RNA. In order to activate the complex, proteins from the Argonauate family are needed. These proteins have two RNA binding domains: a PAZ domain that could bind the single stranded 3' end of the mature miRNA and a PWI domain that interacts with the 5' end of the guide strand. Once they bind the mature miRNA, they orient it towards the target mRNA for interaction (Kim V. Narry, 2004).
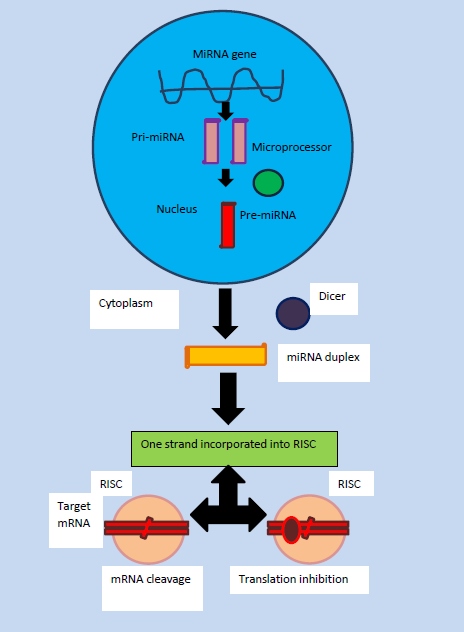
(Marek Mraz, 2004)
Figure 3: Biogenesis
Significance of microRNA in diseases
Although miRNA plays an important role in normal functioning of eukaryotic cells, it is also involved in certain diseases related to the nervous, endocrine, and cardiovascular systems. In the initial development of the nervous system, miRNA assists in the formation of synapses and dendrites. It's presence in the early developmental stages of the heart is essential for normal function. Studies have shown that the expression of miRNA in diseased heart tissue causes the muscles in the heart to weaken resulting in cardiomyopathies. In addition, miRNA exhibits a negative effect on the differentiation of stem cells into adipocytes.
Cancer
Cancer is a complex and devastating disease which necessitates a variety of changes in gene expression and structure. Historically, cancer research has focused on protein-coding genes, considering them to be the primary effectors and regulators of tumorigenesis (Espinosa C. E. et al, 2006). Recent research has established that non-protein coding miRNAs are significantly involved in the regulation of cell growth, differentiation and apoptosis. Although their mechanism of action and their significance in cancer are still at research level, they may be considered important regulators of tumorigenesis genetic expression and thus become important therapeutic tools in the near future. In this way miRNAs acting as possible tumorigenesis regulators function both as tumor suppressors and as oncogenes (Esquela Kerscher A. et al, 2006 ).
miRNAs as tumor suppressors
miR-15a and miR-16-1 are two specific miRNAs, the expression of which was partly or entirely diminished in greater than 65 percent of Chronic lymphocytic leukemia (CCL) cases. The discovery that these two miRNAs and the chromosomal region 13q14 are not found in 50 percent of cases of mantle cell lymphoma, 16 to 40 percent of cases of multiple lymphoma and 60 percent of cases of prostate cancer strongly indicates the function of miR-15a and miR-16 as tumor suppressor genes. Although their full target complement is not yet established, they appear to predominantly mediate their effects by down-regulating the anti-apoptotic protein BCL2. This protein in its 3’UTR transcript exhibits binding sites for miR-15a and miR-16-1 and consequently the co-expression of the latter negatively regulates the BCL2 3’UTR transcript. Although further research is required to prove that the miR-15a and miR-16-1 exert their effects though the BCL2 protein or other additional transcripts in the CCL, these two microRNAs play a significant role in the prevention of lymphomagenesis and leukaemogenesis (Calin GA et al, 2002)
miRNAs as oncogenes
miRNAs can also act as oncogenes either in a direct way by down-regulating tumor suppressors or in an indirect way by down-regulating genes which can act to limit the activity of known oncogenes. An example of this may be observed in the interaction between miR-155 and the MYC oncogene. The latter oncogene is a transcription factor which is able to control the cell growth through the induction of both cell proliferation and apoptosis (Pelengaris S. et al, 2002 ).
(Kusenda B. et al, 2006 )
Figure 4: Role of miRNAs in cancer
Conclusion
miRNA found in both plants and animals functions as a post- transcriptional regulator of gene expression, although with some differences in mechanism of function. The biogenesis of miRNA consists of four steps including: transcription, nuclear processing, nuclear export, and cytoplasmic processing. Once miRNAs have been produced in the body, they induce gene silencing by binding to target sites. This results in a decreased protein activity or the degradation of mRNA. miRNA 's role in cancer, heart and neurological diseases has been outlined.Ongoing research into the function of miRNA in cancer may lead to their use as important therapeutic tools. miRNA have been found to to be involved in cell cycle control, apoptosis, several developmental and physiological processes including stem cell differentiation,embryogenesis, hematopoiesis, cardiac and muscle development, neurogenesis, metabolic regulation and viral resistance. Further research will hopefully continue to provide evidence of the crucial role miRNA plays in eukaryotic organisms.
References for Text
Buckingham, Steven 2003- The Major World of micro RNAs
Mraz, Marek, 2004- MicrRNAs: new regulators of mRNA expression
References for Diagrams
(Recreated by Eirini Stylianou and Caoimhe May Soffer)
Mraz Marek, 2004- MicroRNAs: new regulators of mRNA expression
