Poly (ADP ribose) polymerase (PARP)
Contents
Introduction
The family of Poly (ADP ribose) Polymerase (abbreviated PARP) is a big family of enzymes that have many different tasks in the body (Zaniolo et al, 2007). There are 18 members in the family (Makogon et al, 2010), but our focus will mainly be on PARP-1, the most researched enzyme of this group. It is necessary to clarify that this enzyme has more than one task, and to avoid confusions we will briefly explain the PARP-1 functions to understand further reading. PARP-1 works as a regulatory enzyme in most importantly cell DNA repair and cell death. It will act as a repair enzyme when the damage to the DNA of the cell is small and not too extensive. If, however, the damage to the cells DNA is too extensive, PARP-1 will function as a regulator of apoptosis (the cells programming for self-destruction) and does not repair the DNA damage, but induce cell death. These two main aspects are important to keep in mind for further reading and understanding. PARPs were identified for the first time in 1963, and in 1980 it was discovered that PARP inhibition can enhance DNA damage, a principle which can be used to treat cancer, since the overexpression of PARP-1 is associated with the progression of many types of cancer.
Structure
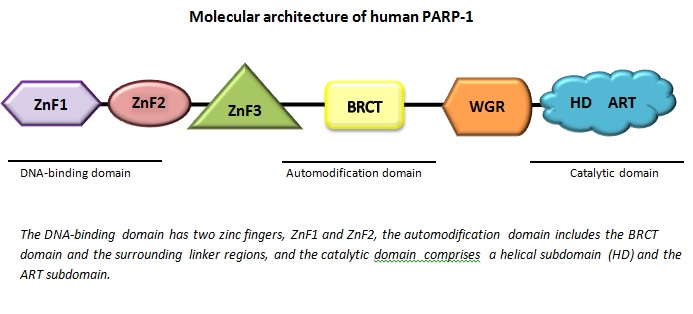
All PARP enzymes consist of different parts called domains. The one domain we can find in all members of the family is the catalytic domain, which forms the active site where the reaction occurs. In addition to the catalytic domain, PARP enzymes also have one or two more domains with various properties. The tasks and functions of the different members of the PARP family depends on which combination of these domains can be found in the particular enzyme.
One of the domains is called zinc finger domain. It’s a double zinc-containing domain for binding of the damaged DNA. One of the zinc molecules will attach to the minor groove of the double helix of the DNA, capturing the free (broken) DNA strand, while the other one will attach to the major groove of the helix. When the zinc has bound to a DNA the enzyme is activated, a central automodification domain will cause structural changes in the enzyme. The automodification domain found in PARP-1 is called BRCT domain (Virág, 2005).
Function
Function of PARPs can be found in almost all eukaryotic cells. PARP-1 is by far the most important member in this family - it is the most abundant one and stands for over 90 % of the actions that this group carries out in the body (Virág, 2005). Although, PARP-2 is not to be forgotten, as it together with PARP-1 is the only member that is activated by DNA damage. PARP-1 is a multifunctional enzyme with many different biological functions. It is located and carries out its actions inside the nucleus of cells. It works as a catalyzer of different molecules that have their effect on proteins within the nucleus, and change their main function (Zaniolo et al, 2007). It carries out its task by catalyzing the addition of ADP-ribose (ADPR) units from nicotinamide adenine dinucleotide (NAD+). This forms a structure called histone, an important and highly modified protein within the cells nucleus (Makogon et al, 2010. Wolfe, 1993, p.542) Several histones together form a nucleosome, which are responsible for the packaging of the DNA. This appearance is often referred to as “beads-on-a-string”, where DNA winds around the nucleosomes. This is very important in the chromatin formation and the activation and inactivation of the genes. (Wolfe, 1993, pp. 549-552)
Other functions of PARP-1 include DNA repair, gene transcription, regulation of protein synthesis, DNA replication, cell differentiation and regulation of cell death. Of these functions we will empathize PARP-1s function in DNA repair and cell death, as these are the main functions of the enzyme.
DNA repair: As mentioned before, PARP-1 has three domains; one responsible for DNA binding, one for automodification, and the last for catalysis. In case of damage to the DNA-strand, PARP-1 will bind to the site of damage through its double zinc finger DNA binding domain. This binding will result in the activation of the enzymatic activity of PARP-1 (Kim et al, 2005) and there will be a formation of long so-called PAR chains. These long molecular chains are negatively charge and will attract DNA repair proteins to the site of damage. It will also remove PARP-1 from this area so that more repair proteins can access.(Kummar et al, 2012)
Cell death: In the presence of high level of DNA damage, PARP-1 will no longer promote DNA repair but have a contrary role; promoting cell death. A severe DNA damage will result in high production of PAR chains that can cause death of the cell in two ways. It can either induce necrosis, which is a process where the cell dies because of abnormal swelling and that eventually leads to the rupture of the cell. Another type of cell death is apoptosis. This is a highly organized process and the cell is completely destroyed. The most important trigger of apoptosis is a protein called AIF, and PARP-1 can play a role in the apoptosis through this protein. Exact how PARP-1 promote cell death through both necrosis and AIF is not fully understood (Kim et al, 2005). For cell death to happen through apoptosis, there has to be sufficient amounts of ATP available. Cells canloosetheir ATP pool due to PARP-1 activation, which leads to that these cells may die through necrosis instead (Kim et al, 2005).
Regulation of PARP-1
There have been many studies during the last decade in research related to the function of PARP and which inhibitors and agonists that can be found. According to recent research production of PARP-1 have been shown to be related to cell growth, and not by DNA-damage (Kummar et al, 2012. Zaniolo et al, 2007). There have been found different promoters of the gene expression of the PARP-1 enzyme, some working as activators for transcription of the enzyme, while others can work as repressors and down regulating production of the enzyme. PARP-1 itself also contributes as a stimulator or a repressor. This means the enzyme itself has a dual self-regulatory function of gene transcription (Zaniolo et al, 2007).
Activation of the PARP-1 enzymatic activity
Whilst the main production of PARP-1 is initiated by cell growth, such as growth of tumors, there are several ways to fully activate the enzyme (Kummar et al, 2012). The main stimulator is the breaks or damage of DNA, which usually occur due to environmental stimuli and free radicals. PARP-1 is able to bind to damaged DNA in the forms of both single-strand breaks and double-strand breaks. Are there low levels of damage and breaks, the PARP-1 will function as a repair-enzyme, while if there is high damage and a lot of breaks PARP-1 will induce apoptosis, controlled cell death. The basal enzymatic activity of PARP-1 is very limited, but it is activated and stimulated to a high degree in the presence of a variety of cofactors such as damaged DNA, some undamaged DNA structures, nucleosomes and different protein-binding partners (Kummar et al, 2012).
Inhibition of the PARP-1 enzymatic activity
Compared with other pathways of PARP-1, it has been shown that the effect of inhibiting PARP-1 is especially successful if the tumor already has defects in its DNA damage repair pathways (Kummar et al, 2011). Research conclude that inhibition of PARP-1 enzyme as for now have little or no effect on other DNA repair pathways, and it remains a question if PARP-1 inhibition will be successful with tumors without defects or in other DNA-repair pathways (Kummar et al, 2011).
PARP-1 inhibitor that has been clinically developed, shows that they will bind to the catalytic domain of the enzyme and therefore inhibit automodification. Also, the release of the enzyme from the DNA damage site inhibitis other repair proteins to the site of the DNA damage, not resulting in DNA repair. The goal of inhibiting cell DNA repair of cancerous tumor cells in combination with common methods for cancer treatment, such as cytotoxic chemotherapy and radiation technology, will lead to greater success in removing and treating the cancer (Kummar et al, 2011). As for today, cytotoxic chemotherapy is used on its own treating cancer, and with new research on PARP-1 in combination with the regular treatments may revolutionize the cancer treatment therapy. The clinical trials of PARP-1 inhibitors are in progress, and some of the inhibitors discovered so far are Rucaparib, Iniparic, Olaparib, Veliparib and other inhibitors. These are in the process of testing, some with animal experiments, others with human volunteers in clinical trials (Kummar et al, 2011).
PARP in the healthy cell
Cancer develops when there is instability in the chromosomes found within the cell nucleus, leading to a faulty replication of the genetic material. The instability of the chromosomes is often a result of earlier faulty DNA-repair or defect cell cycle.
In a healthy cell, PARP is one of several cancer suppressing factors with responsibility to stabilize the chromosomes, and therefore prevent cancerous growth (Kummar et al, 2012. Patel and Kaufmann, 2010). The importance of PARP in healthy cells has been proven with animal experiments with so-called knockout mice. A knockout mouse has a genetically modified DNA where some genes have been selectively removed. In this experiment the gene coding for PARP have been absent or present, parallel with modification of a gene for another cancer suppressing protein, p53. Studies of the p53 have shown that it has a role in the cell cycle and certain checkpoint in the apoptosis. Cells lacking this protein have been shown more susceptible of cancerous development.
The p53 and PARP have a close interaction in mammalian cells. Studies have shown that PARP can bind to specific domains of the p53 and modify its activity. Absence of PARP will not only decrease the abundance of the p53, but also affect the efficiency when DNA-damage occurs. In the mice where the PARP was present but not the p53, the mice rarely developed carcinomas (a cancerous group of tumors). In mice where the PARP gene was missing but p53 present, the mice showed a dramatic increase of tumors of carcinoma type (Kummar et al, 2012).
This outcome shows that PARP most likely is important in interaction with other molecules supporting and protecting the genetic material. The loss of PARP will also result in the loss of other tumor suppressing functions, leading to a cancerous development (Kummar et al, 2012).
As the other main function of PARP is cell DNA repair, PARP can also function as a helper for cancerous growthin cells where abnormal development already begun. This can be explained with the fact that if the damage of the DNA is not extensive, the PARP will repair the cell, thus repairing the cancerous formation and assist continued growth and developing of the tumor (Zaniolo et al, 2007).
PARP-1 inhibition is only related to those tumor cells where PARP-1 will function as a repair enzyme. With inhibition of the PARP-1 in these cells, they will be easier treated with cytotoxic chemotherapy, as it has been shown that these treatments have greater success with cells that have DNA damage (Kummar et al, 2012). This decreases the side effects, such as killing surrounding healthy tissue, which is a problematic effect in today’s cancer treatment (Kummar et al, 2012).
Discussion
Will inhibiting PARP-1 simultaneously with treating the patient with cytotoxic chemotherapy, such as those used in treatment of cancer, enhance the overall treatment success with little or no side effects? The diagnosis of cancer is closely related to overexpression of PARP-1, especially in breast cancer, and therefore research on inhibition has been and still is highly developing. The scientists now hope to be able to use PARP technology as a complement to other, or even replacement, for cancer treatments such as radiation or cytotoxic chemotherapy (Koziol et al, 2012). The benefits of PARP would be a treatment that acts more specific, and not general, destroying and damaging other non-cancerous cells in the body. Clinical trials have begun, but have unfortunately been limited. When traditional chemotherapeutic agents have been reduced and partially replaced by PARP inhibitors, side effects such as decreased amounts cells responsible for immunity, oxygen transportation and blood clotting (myelosuppression) have been seen (Patel and Kaufmann, 2010).
Photo of a dog operated for mammary carcinomas (Lindberg, 2011)
References
Litterature
Wolfe, Stephen L. (1993). Molecular and cellular biology. University of Michigan, Wadsworth Pub. Co. pp. 542, 549-552
Pictures
Malisius, J. K. L. (2012): Molecular architechture of human PARP1, redrawn based on the figure of http://www.nature.com/nsmb/journal/v19/n7/images/nsmb.2306-F1.jpg
Lindberg, Birger (2011): Photo of a dog operated for mammary carinomas
